Each year, pharmaceuticals, biotech organizations, academic institutions and law firms commit costly errors that happen due to poorly informed IP portfolio decisions. Relating to biological sequence search, here are nine serious mistakes we hate to see life science companies make.
1. Overlooking patent sequence data.
Serious sequence information search require specify and organized efforts, and searching Genbank is not enough. Genbank had 180 million sequences as of its December 2014 build, only 32 million of which are identified in their patent division. As a contrast, GenomeQuest’s GQ-Pat had over 280 million sequences, all found in patents, almost nine times larger.
2. Under-utilizing annotation information.
Ascertaining the legal or biological importance of the similarity between any two sequences requires a clean, curated database with organized annotation fields and content. Additional fields, such as bibliographic references, date of earliest publication, and date of sequence disclosure add analytical speed and precision when used with a rapid search result filtering function.
3. Forgetting the Dark Genome.
4. Taking too much time.
5. Hoping for the best.
Moving forward with a research project without properly searching and evaluating sequences can prove to be costly in the long run. An incomplete evaluation of the data early in the research cycle can be costly once a completed project is found to have yielded unusable results.
6. Making decisions based on yesterday’s results.
Genome sequence information is extremely dynamic. In addition to the steady addition of recorded primary sequence data, scientific and patent information about both new and previously existing sequences also grows and changes on a daily basis. A sequence data query affecting important scientific research and business decisions might not yield the same answer one week from now. The more sequences involved in the decision, the greater the risk. Research groups and businesses without access to an automa
ted and continuous search-and-report system are particularly vulnerable.
7. Using the wrong algorithm.
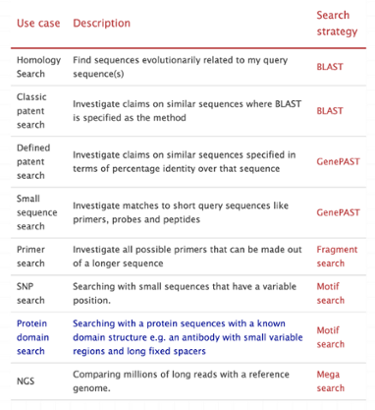
Even the most experienced analyzers can make a mistake choosing the right algorithm for sequence search. For example, using BLAST for short sequences will miss many approximate hits. GenePAST is a better algorithm to use in many sequence search cases.
8. Too many gatekeepers.
Restricted access rights to proprietary databases, cumbersome search software user interfaces, and outdated business practices often prohibit direct utilization of sequence data search systems by the person asking the question, who must instead work through one or more gatekeepers. A well-defined project submission process can prevent intended queries from getting “lost in translation,” but when sequence searches are outsourced, queries are often composed broadly in order to prevent potentially relevant results from being excluded from the search report returned by the service. This results in an oversized report and a long manual search process for the sequence records of real interest. Gatekeeper delays also inhibit creative sequence data exploration, where hunches and hypotheses can be quickly formed and investigated using fast, iterative database queries.
9. Ignoring workflow issues.
Commercially licensed or in-house bioinformatics solutions often become very popular within organizations as researchers learn to use them to great advantage. But an effort to provide genome search capability to the user base that does not consider workflow issues can result in the installation of an isolated, standalone information “silo” with an unfamiliar interface. The standalone solution is itself likely to be underutilized, and also fails to take advantage of organizational knowledge built up around previously existing bioinformatics applications.